Biography
Bachelor in Biotechnology, Igor Sikorsky Kyiv Polytechnic Institute, Ukraine.
Masters of Chemoinformatics, Institute of High Technologies KNU, Ukraine.
Intern in MDC, Rajewsky Lab. Project: Single-cell analysis of human brain organoids.
Secondments
Secondment 1: Optimization (efficiency and accuracy) of semantic multi-omics data integration pipelines
Secondment 2: Review of available mouse model experimental results and public catalogues to set up experiments to validate the findings
ESR 12 Project
Multi-omics analysis to delineate drug-response pathways
The advent of high-throughput techniques provided petabyte amounts of heterogeneous biomedical data such as genomics, transcriptomics, proteomics, metabolomics, and other omics, which together could be used to achieve personalized and precise medicine and also upgrade current biomedical research by making it more translative. However, there are still several challenges in multi-omics data integration and interpretation, such as noise, difference in scale, heterogeneity, lack of normalisation, etc. Therefore development of new data science techniques for omics data processing is needed.
One of the key opportunities of multi-omics-based personalised medicine is to precisely predict individual drug responses and find the most efficient treatment approach by systematic analysis of patient’s omics data. This could be done by development of a representative prediction model or applying algorithms, which integrates a patient’s genome, exprosome, biomarkers range and clinical data with possible treatment approaches. Particular interest belongs to single-cell multiomics and development cell-type specific gene regulatory network pipelines.
The main challenges in multi-omics data processing are heterogeneity and sparsity of data, its size (difficulties to process large datasets) and dimensionality. In our research we are going to validate and implement existing tools such as single-cell processing pipelines and multi-omics algorithms, and develop custom scripts and software.
My research group has very recently observed that such regulatory networks differ between individuals, and thus are personalized (Van der Wijst, Nature Genetics 2018 and Van der Wijst, Genome Medicine 2019). This has immediate ramifications for the efficacy of drugs. E.g. in this figure two individuals (John and Kate) are denoted.
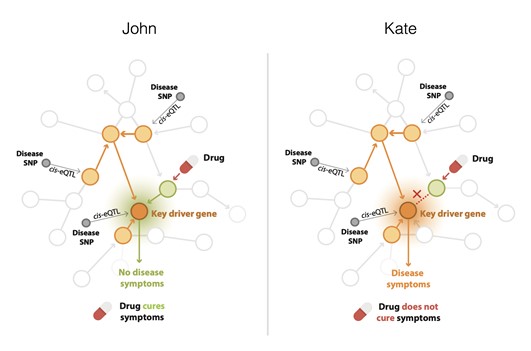
Due to the existence of a regulatory relationship between the green gene and orange gene for John, the drug, acting on the green gene is effective in resolving disease symptoms.
However, due the absence of that relationship for Kate this particular is not going to be effective for her. Therefore, our main focus is analysing personalised cell-type specific gene regulatory networks and their advancement by integrating other omics information. After development, validation and implementation of these individual multi-omics models, we will match them with drug datasets and processes in order to delineate drug response pathways and find personalised treatment scheme.
Overall, the rapid development of bioanalytical techniques provided a multiple large scale datasets, but still their proper and comprehensive processing tools are needed. Moreover, multi-omics data integration could substantially contribute to understanding disease progression and individualized drug influence, which is the main aim of this research.
Scientific Interests
-
Single-cell multiomics
-
P4 Medicine
-
Data integration
General Interests
-
-
Biomedical startups and innovative business
-
Writing





