
The TranSYS ITN holds a monthly journal club, organised by the TranSYS research council for its 15 ESRs. Each month a scientific paper is selected by majority from a list of three interesting papers and an ESR presents followed by a discussion with ESRs and PIs.
TranSYS is a highly multidisciplinary network and the Journal Club brings a perfect opportunity to learn about other participants’ domains and interests, as the paper presented may be potentially applied to our own project or at least shed some insight into other areas of research.
As this month’s presenter, I chose to talk about a method paper, entitled “Unsupervised discovery of phenotype-specific multi-omics networks”, published on Bioinformatics in 2019. The authors developed a pipeline to build sparse CCA-based multi-omics networks that are specific to a 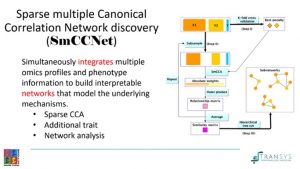 quantitative phenotype. CCA has been widely used as a multivariate method for data integration. It finds the optimal (linear) transformations of the 2 data modalities so that they’re maximally correlated. In case of too many features or features with little contribution, sparsity was introduced. And to account for the additional phenotype, a weighted form of multiple CCA was applied on top of the sparse CCA. The canonical weights coming out of the CCA-based approach were then used to generate the similarity matrix on which a hierarchical tree cut was performed to derive the most coherent modules specific to the phenotype.
quantitative phenotype. CCA has been widely used as a multivariate method for data integration. It finds the optimal (linear) transformations of the 2 data modalities so that they’re maximally correlated. In case of too many features or features with little contribution, sparsity was introduced. And to account for the additional phenotype, a weighted form of multiple CCA was applied on top of the sparse CCA. The canonical weights coming out of the CCA-based approach were then used to generate the similarity matrix on which a hierarchical tree cut was performed to derive the most coherent modules specific to the phenotype.
This paper would be interesting to anyone who works with multiple data types, e.g. multi-omics data, and wants to summarize the features from both datasets with a particular interest in an additional phenotype. After my presentation, several ESRs raised questions about both technical details and general applications of this method. It was an informative and stimulating discussion that not only clarified the ambiguity in the complex pipeline but allowed ESRs to think about how their own projects may benefit from this presentation.
Reference: W Jenny Shi, Yonghua Zhuang, et al. Unsupervised discovery of phenotype-specific multi-omics networks, Bioinformatics, Volume 35, Issue 21, 1 November 2019, Pages 4336–4343, https://doi.org/10.1093/bioinformatics/btz226

Author: Zuqi Li
TranSYS ESR 2: Hunting for patient subtypes through image-based phenotypes as biomarkers for major gene effects in medical disorders


